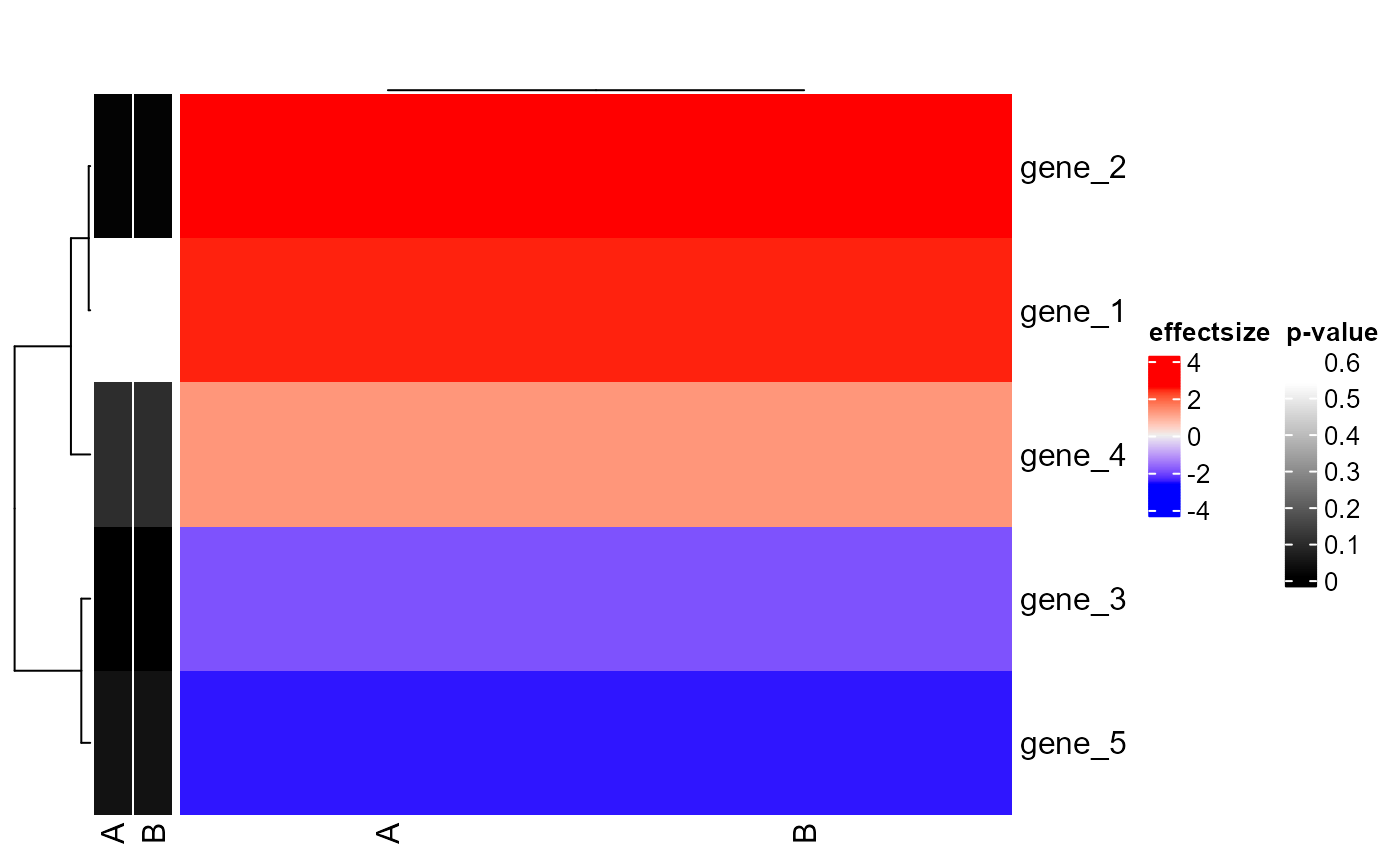
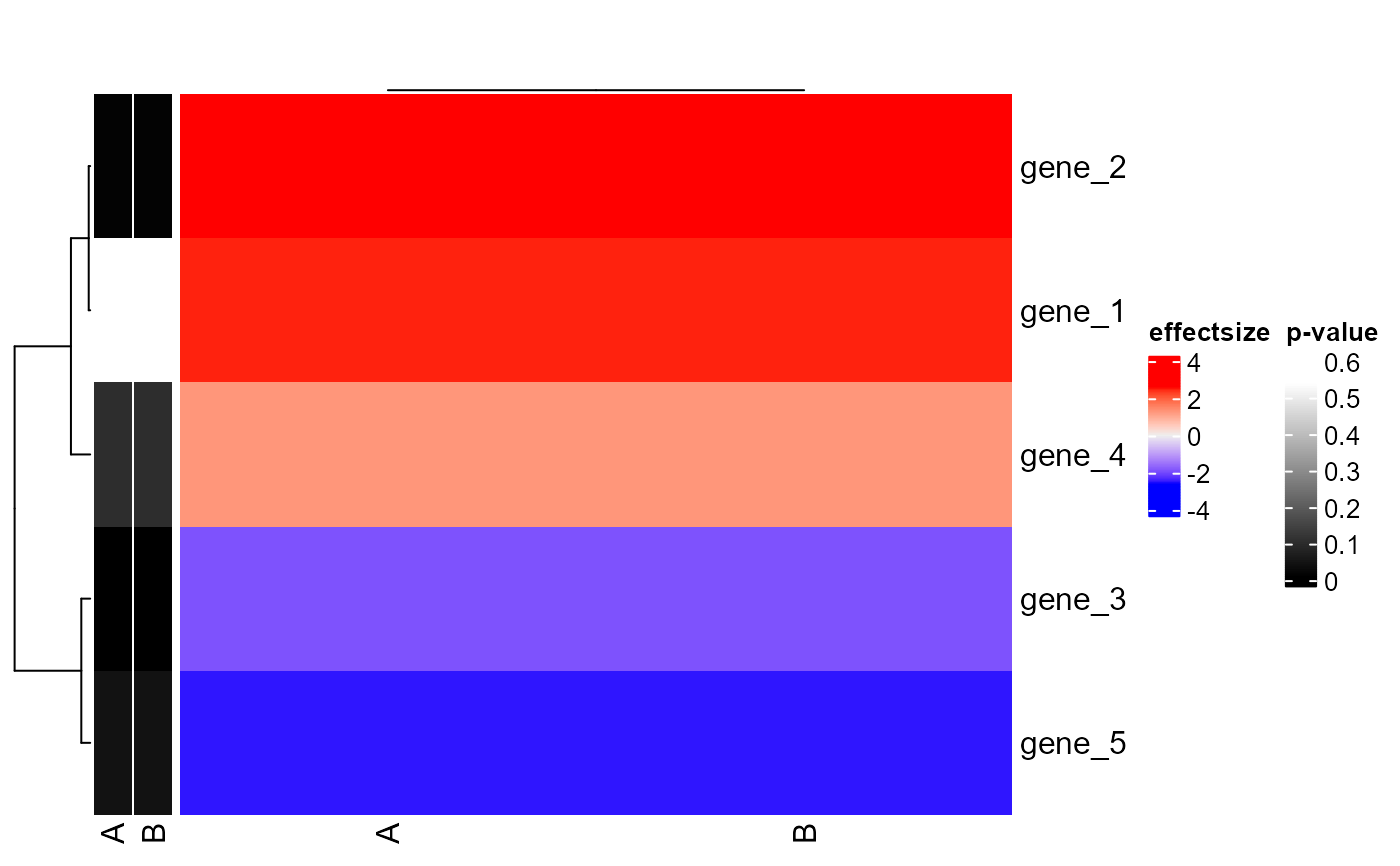
Plot gene2effectsize ComplexHeatmap
Source:R/plot_gene_effectsize_ComplexHeatmap.R
plot_gene_effectsize_ComplexHeatmap.RdPlot gene2effectsize ComplexHeatmap
plot_gene_effectsize_ComplexHeatmap(
genes,
genes_overview,
rows_dendrogram = TRUE,
cols_dendrogram = TRUE,
plot_n_genes = 50
)Arguments
- genes
character, genes to visualize
- genes_overview
dataframe, containing columns: 'symbol', 'SAMPLE_efsi' and 'SAMPLE_pval'
- rows_dendrogram
default: FALSE, TRUE to cluster rows and show dendrogram
- cols_dendrogram
default: FALSE, TRUE to cluster columns and show dendrogram
- plot_n_genes
integer, default: 50, NULL to plot all genes
Value
ComplexHeatmap object
Examples
plot_gene_effectsize_ComplexHeatmap(
c('gene_1', 'gene_2', 'gene_3', 'gene_4', 'gene_5'),
get(load(system.file("extdata", "example_genes_overview.rda", package = "goatea")))
)